 Predict the effective gRNA sequences.
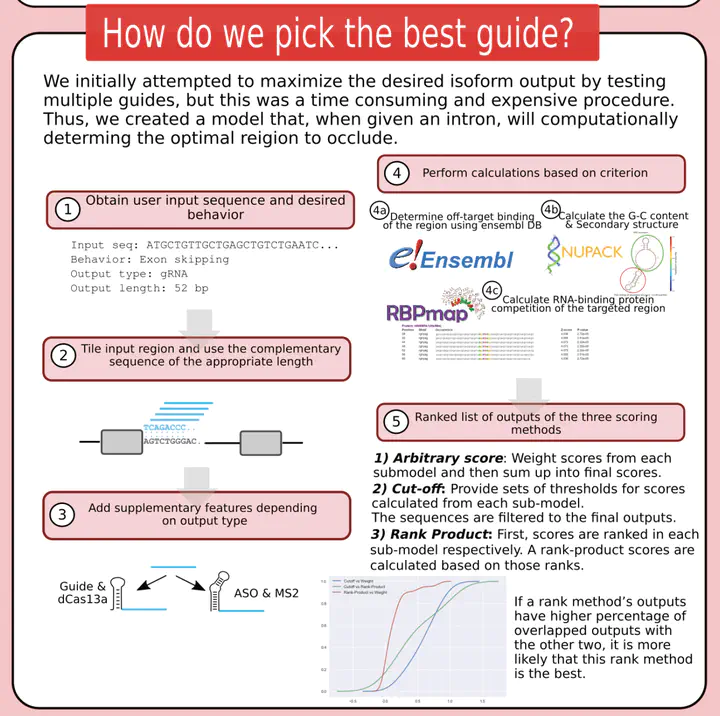
Predict the effective gRNA sequences.Alternative splicing is the eukaryotic mechanism that selects which exons will be included in processed mRNA. It increases the diversity of the human proteome by allowing more than one protein isoform to be produced from the same gene. The inclusion or exclusion of particular exons is regulated by splicing factors which bind to specific motifs in the pre-mRNA. Our project aims to control alternative splicing by using the RNA-binding proteins dCas13a and Ms2 to interfere with these splicing factors’ binding, allowing us to control which exons are included in the final processed mRNA. By choosing between different protein isoforms, this control method may allow synthetic genes to dynamically change functionality. It also has therapeutic potential for diseases that arise from aberrant splicing, including an aggressive form of breast cancer where one splice variant produces a nonfunctional tumor suppressor, contributing to tumor formation. My work focuses on building a simple computational model that predicts the effective gRNA sequences to target introns that we desire to bind and cause exon skipping. The model considers four factors - GC Content, secondary structure, off-target mismatches and RBP competitions. Our team work can be found on the iGEM website.